|
Clara Wong-Fannjiang I'm a Senior Machine Learning Scientist at Genentech in South San Francisco, where I am part of the Frontier Research team within Prescient Design. My research works toward facilitating trustworthy AI-guided scientific inquiry and decision-making, with a current focus on quantifying and managing the consequences of uncertainty in AI predictions. I received my Ph.D. in Electrical Engineering and Computer Sciences from UC Berkeley in August 2023, where I was advised by Michael I. Jordan and Jennifer Listgarten as an NSF Graduate Research Fellow. I also spent a summer working with Nikhil Naik and Ali Madani at Salesforce Research. Prior to Berkeley, I conducted marine biology fieldwork with Kakani Katija at the Monterey Bay Aquarium Research Insitute and worked with David Sussillo as a Google AI Resident. I received my B.S. in Computer Science in 2016 from Stanford University, where I benefited from the guidance of Stephen Boyd, Coraline Rinn Iordan, and Fei Fei Li. Beyond research, I am an avid hiker, fan of women's artistic gymnastics and aroids, and proud mom to an extraordinary son. |

|
ResearchI am broadly interested in the question of how to make trustworthy machine learning-guided scientific conclusions and decisions. My current work tackles this question through the lens of uncertainty quantification methods, and is particularly motivated by problems in drug discovery and clinical decision-making. Some highlighted work is below, where (α-β) denotes alphabetical ordering and * denotes equal contribution. |
|
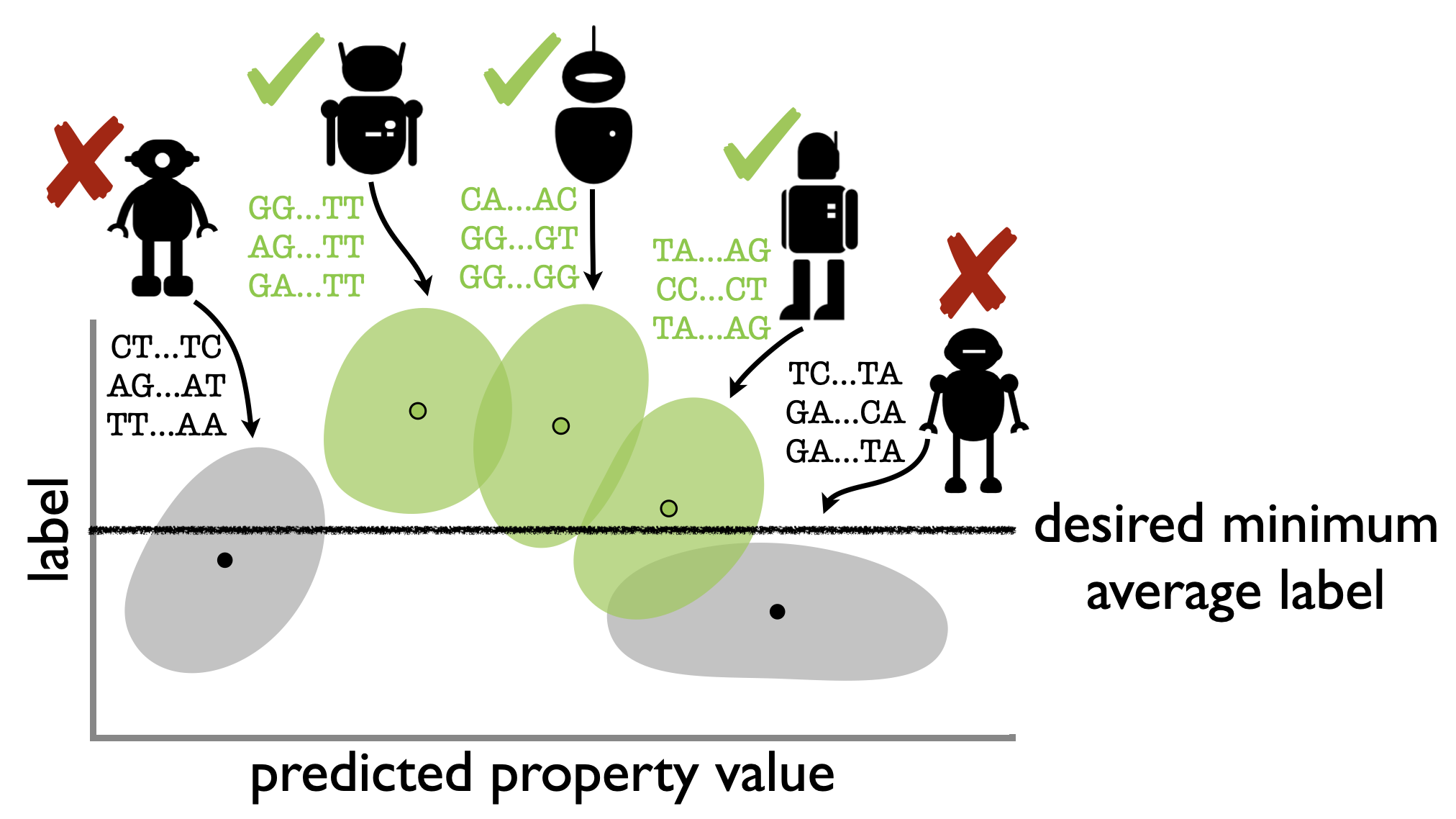
Reliable algorithm selection for machine learning-guided design
Clara Fannjiang and Ji Won Park International Conference on Machine Learning 2025, Machine Learning in Computational Biology 2025 (oral) ICML (open access) · arXiv · code · BibTeX How can we anticipate whether an algorithm will be successful for a machine learning-guided design task? We use prediction-powered inference techniques to reliably forecast characteristics of the distribution of designs that will be produced by an algorithm, enabling the selection of algorithms that will be successful (if there are any) with high-probability guarantees. |
|
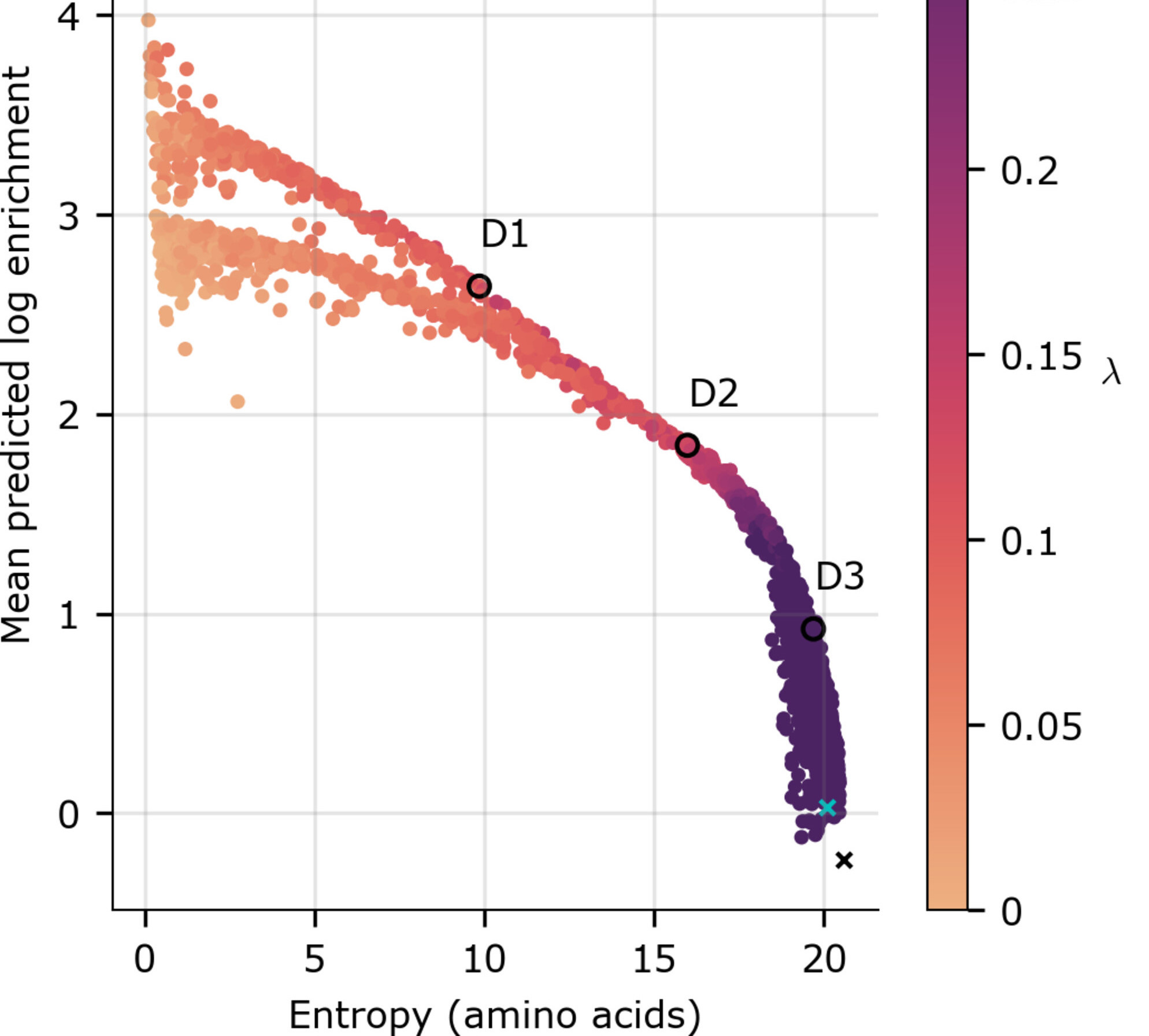
Optimal trade-off control in machine learning–based library design, with application to adeno-associated virus (AAV) for gene therapy
Danqing Zhu*, David H. Brookes*, Akosua Busia*, Ana Carneiro, Clara Fannjiang, Galina Popova, David Shin, Kevin C. Donohue, Li F. Lin, Zachary M. Miller, Evan R. Williams, Edward F. Chang, Tomasz J. Nowakowski, Jennifer Listgarten, and David V. Schaffer Science Advances, 2024 Sci. Adv. (open access) · bioRxiv · BibTeX A diverse set of designed objects enables a practitioner to hedge against unknown future desiderata. We develop a machine learning-guided design method that navigates a trade-off between design diversity and achieving desirable predicted values, and apply it to the design of diverse AAV capsids with high packaging efficiency. |
|
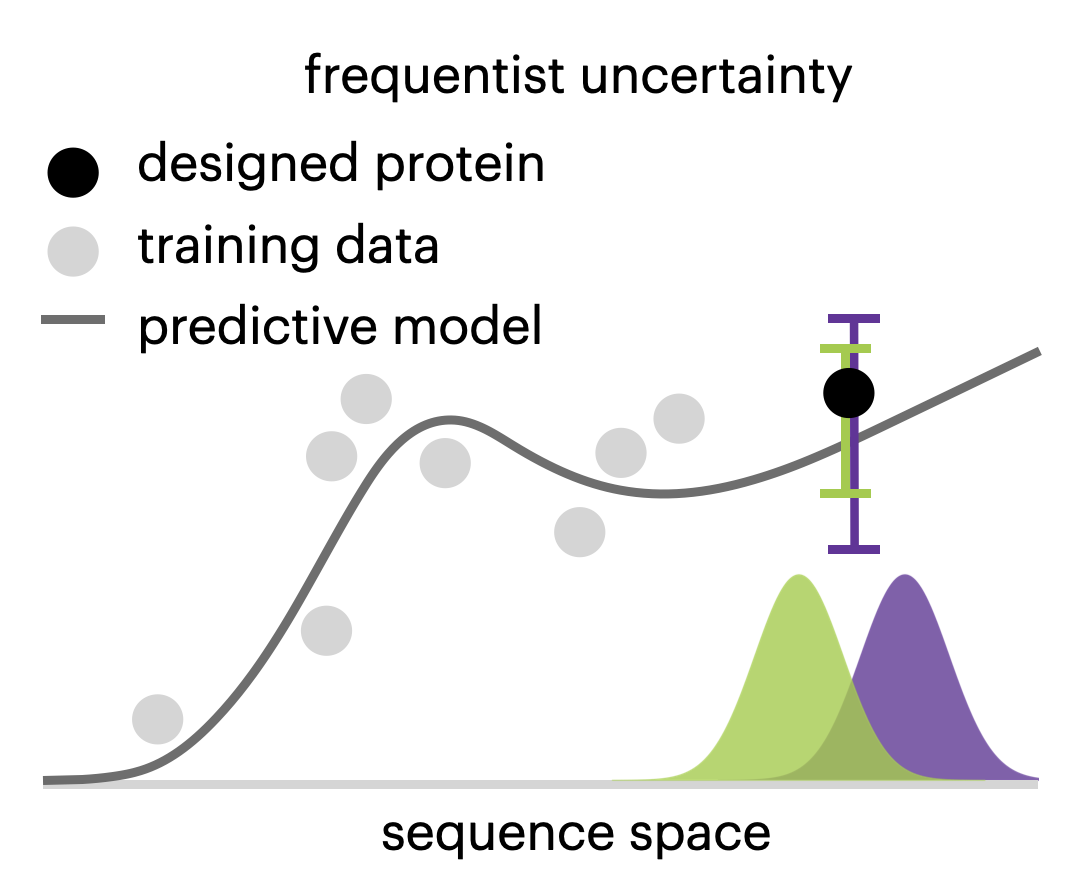
Is novelty predictable?
Clara Fannjiang and Jennifer Listgarten Cold Spring Harbor Perspectives in Biology, 2023 CSHL · PDF · arXiv · BibTeX A discussion of considerations when using machine learning-based predictions to guide the design of novel objects, which by definition differ from the training data and are therefore particularly prone to prediction error. |
|
Prediction-powered inference
(α-β) Anastasios N. Angelopoulos, Stephen Bates, Clara Fannjiang, Michael I. Jordan, and Tijana Zrnic Science, 2023 Science · PDF · arXiv · code · BibTeX We develop a framework for performing valid statistical inference—constructing confidence sets and p-values—when gold-standard data is supplemented with predictions from any predictor, machine learning-based or otherwise. |
|
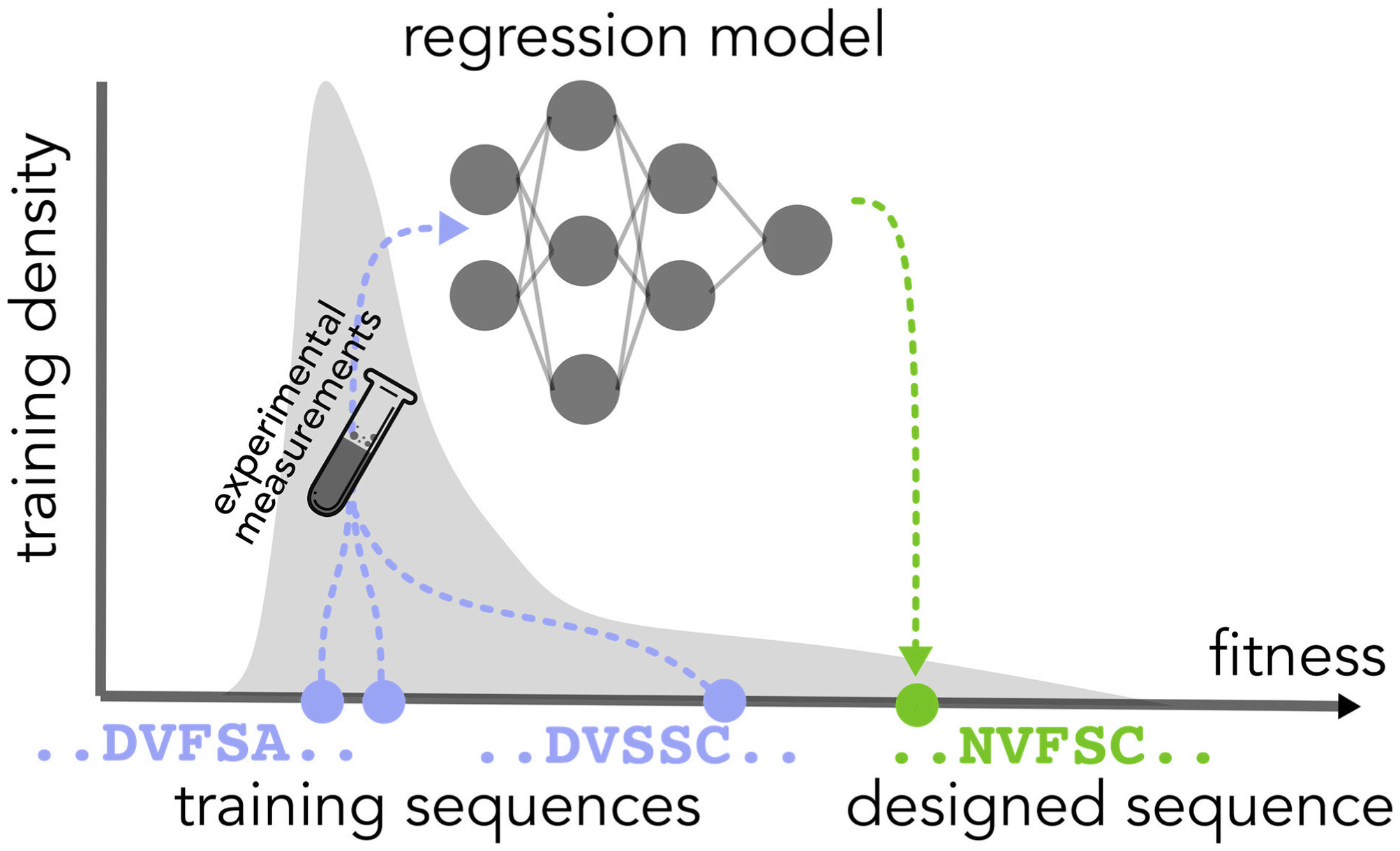
Conformal prediction under feedback covariate shift for biomolecular design
Clara Fannjiang, Stephen Bates, Anastasios N. Angelopoulos, Jennifer Listgarten, and Michael I. Jordan Proceedings of the National Academy of Science, 2022 PNAS (open access) · arXiv · code · BibTeX We characterize feedback covariate shift, a type of distribution shift in which the test and training data are dependent: the former is determined by the latter, such as in machine learning-guided design. We generalize conformal prediction to the feedback covariate shift setting. |
|
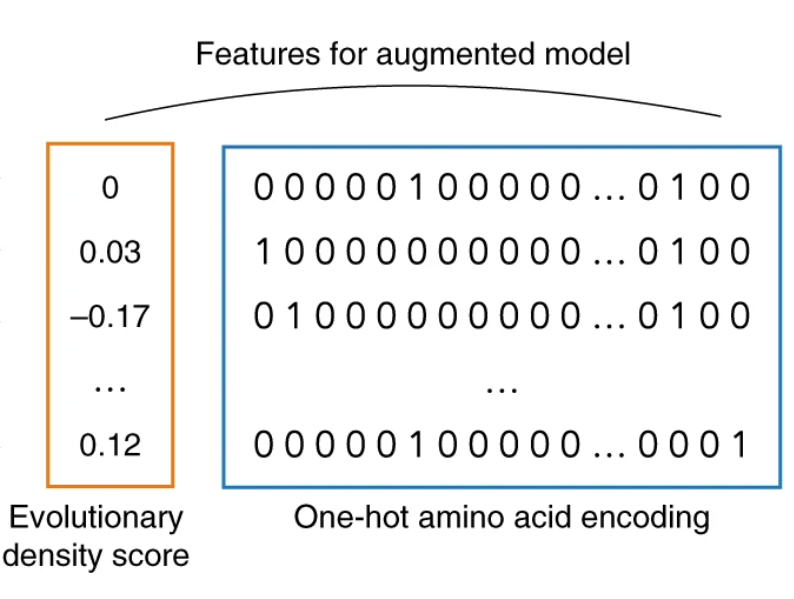
Learning protein fitness models from evolutionary and assay-labelled data
Chloe Hsu, Hunter Nisonoff, Clara Fannjiang and Jennifer Listgarten Nature Biotechnology, 2022 Nat. Biotech. · PDF · bioRxiv · code · BibTeX Simple baselines outperform fine-tuned protein language models on protein fitness prediction. |
|
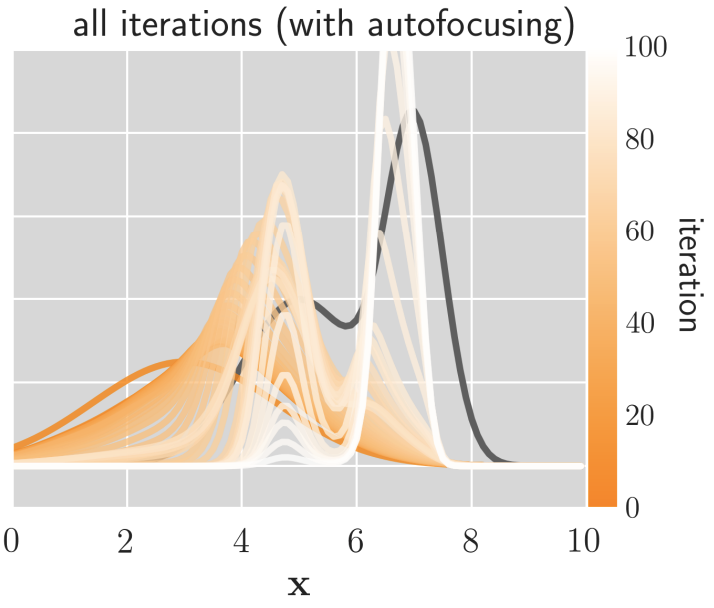
Autofocused oracles for model-based design
Clara Fannjiang and Jennifer Listgarten Neural Information Processing Systems 2020 NeurIPS (open access) · arXiv · code · BibTeX Given fixed training data, algorithms for machine learning-guided design often consult a fixed predictive model. We propose instead retraining the predictive model as such algorithms proceed, on the same training data but carefully reweighted to reflect where the algorithm is currently "focused" in input space. |
|
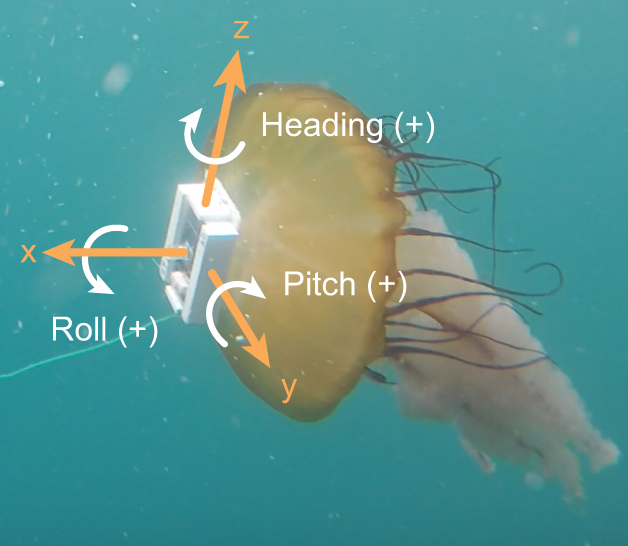
Augmenting biologging with supervised machine learning to study in situ behavior of the medusa Chrysaora fuscescens
Clara Fannjiang, T. Aran Mooney, Seth Cones, David Mann, K. Alex Shorter, and Kakani Katija Journal of Experimental Biology, 2019 JEB (open access) · jellyfish footage · BibTeX A spunky first step toward using machine learning to make sense of wild jellyfish telemetry collected in the Monterey Bay. |
Select Talks
Simons Institute Workshop on AI≡Science: Strengthening the Bond Between the Sciences and AI, June 14, 2024 (recorded).
|
Writing |

|
Monarch McFly: Back to the Evolutionary Future
Clara Fannjiang Berkeley Science Review, Spring 2020 BSR · PDF Pop science piece on work by Karageorgi et al. (2019). Artwork by Alison Nguyen. |

|
Filling in the Species Gap
Clara Fannjiang Berkeley Science Review, Fall 2019 BSR · PDF Pop science piece on Sara A. Stoudt's and Kelly Iknayan's work in statistical modeling in ecology. Artwork by Alison Nguyen. |
|
Updated June 2025. Grateful user of Jon Barron's website code. |